Contact

Laura Hug, Department of Biology, University of Waterloo

Veronica Viljakainen, Department of Biology, University of Waterloo
Introduction
Global concerns with environmental plastic contamination and climate change have led to an increased demand for sustainable alternatives to petroleum-based plastics such as polyhydroxyalkanoates (PHAs). PHAs are made by microorganisms from renewable carbon resources and biodegrade into non-harmful products in natural environments through the action of microbially produced PHA degrading enzymes or depolymerases. With PHA production levels expected to increase, assessing the distribution and diversity of PHA depolymerases is critical for understanding the potential impact of PHAs on the environment and to identify biomolecular tools useful for processing these plastics.
This study used large amounts of publicly available environmental sequence data to characterize the environmental distribution and diversity of PHA depolymerases and to predict novel PHA-degrading enzymes. Specific study objectives were to expand the recognized diversity of PHA depolymerases, inform understanding of the frequency of PHA depolymerases different global environments and identify enzymes and organisms that may prove suitable for future bioremediation, chemical processing or biotechnological applications.
Methodology
A reference set of biochemically validated ePHA depolymerases, the key PHA degrading enzymes, were gathered from the literature and used as queries in a BLASTp search against 3,078 assembled and annotated metagenomes and 5290 metagenome-assembled genomes (MAGs) that were publicly available on the Integrated Microbial Genomes and Microbiomes system. A series of curation steps were taken to increase PHA depolymerase prediction accuracy and to reduce or eliminate non-functional homologues.
All metagenomic datasets had environmental metadata available consistent with the GOLD five-level ecosystem classification system: 1) ecosystem, 2) ecosystem category, 3) ecosystem type, 4) ecosystem subtype and 5) specific ecosystem. To account for differing dataset sizes, frequencies (hits/Gb) for different GOLD level ecosystem classifications were derived by dividing the total number of enzymes identified in metagenomes from an ecosystem category or subtype by the sum total of the sequence data screened from metagenomes occurring within a group. Frequencies in the MAGs for different phyla were calculated by dividing the total number of enzymes identified from a phylum by the sum total of the sequence data screened from MAGs of that phylum.
Phylogenetic affiliations of PHA depolymerases were predicted for the unbinned PHA depolymerase-encoding scaffolds using a diamond-blast search against NCBI's non-redundant database and subsequently conducting a lowest common ancestor analysis (LCA) with MEGAN6 v6.19.5 using GTDB taxonomy.
Outcomes
Study results indicated that PHA depolymerases are found globally and are unevenly distributed across most ecosystems. A total of 13,869 PHA depolymerases were predicted in 1,295 metagenomes from aquatic (28% prevalence in these datasets), solid waste (68%), terrestrial (58%) and wastewater (40%) environments. The highest frequency of predicted PHA depolymerases occurred in wastewater systems (10.73 hits/Gb) and terrestrial environments (8.22 hits/Gb), whereas solid waste and aquatic systems had averages of 5.53 and 4.09 hits/Gb respectively. Wastewater metagenomes, particularly those from activated sludge and nutrient removal systems, had the highest frequency of predicted PHA depolymerases and may be ecological hotspots for these enzymes (Figure 1).
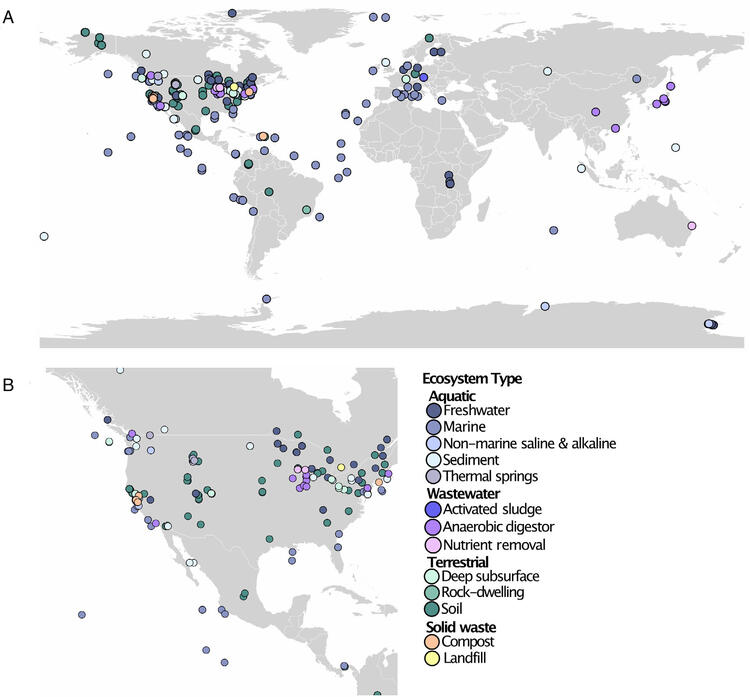
Figure 1: Location (A. World; B. North America) of samples with metagenomes containing one or more predicted PHA depolymerase coloured by GOLD ecosystem.
The study identified medium-chain length PHA (PHAmcl) depolymerase-encoding MAGs from the Proteobacteria, Firmicutes and Myxococcota, with the highest overall frequency occurring in the Firmicutes. Interestingly, a few ePHAmcl-encoding scaffolds were classified as Acidobacteriota, Deinococcota and Dormibacterota, which are lineages that have not yet been associated with these depolymerases. Across ecosystem types, the Proteobacteria (Alpha and Gamma) were the most abundant predicted PHA depolymerase hosts with soil environments having the highest diversity. The Myxococcota-encoded and Chloroflexota-associated PHA depolymerases appeared in relatively high levels in thermal springs and otherwise were only found in soil. Five phyla containing predicted PHA depolymerases were detected in all wastewater ecosystem types - the Proteobacteria, Bacteroidota, Gemmatimonadota, Myxococcota and Spirochaetota. (Figure 2)
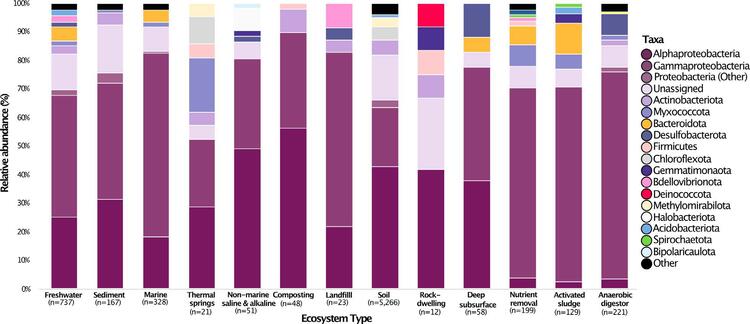
Figure 2: Phylogenetic affiliation of scaffolds encoding at least one predicted PHA depolymerase across different ecosystems, presented as relative abundances. Number of PHA depolymerase-encoding scaffolds for each environmental category is listed (n).
Prior to the study, bacterial ePHA depolymerases were characterized from the Proteobacteria, Actinobacteriota, Bdellovibrionota and Firmicutes, as well as one fungal phylum, the Ascomycetes. This study infers PHA depolymerases occur in 23 additional bacterial phyla and two archaeal phyla. An LCA analysis associated this predicted function with 28 different phylum-level lineages, 10 of which were also identified in the MAG screen. Together, the analyses affiliated predicted PHA depolymerases predominantly with the Proteobacteria, Bdellovibrionota, Myxococcota, Methylomirabilota, Actinobacteriota and Chloroflexota (Figure 3).
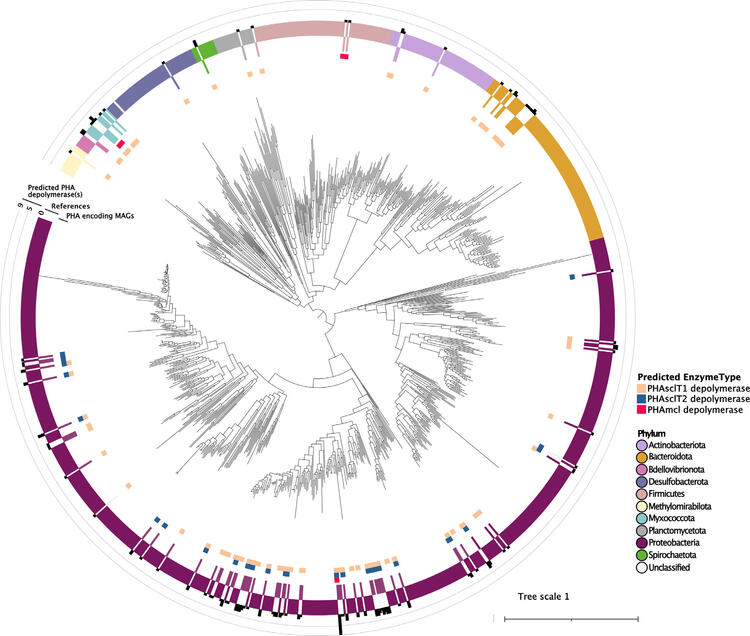
Figure 3: The predicted phylogenetic distribution of MAGs encoding predicted PHA depolymerases. A concatenated 16 ribosomal protein tree inferred using FastTree shows the placement of 98 high-quality, dereplicated MAGs encoding predicted PHA depolymerases placed among reference sequences selected from the bacterial domain. Tree scale bar indicates number of substitutions per site. The coloured ring labelled ‘PHA-encoding MAGs’ is coloured to indicate the phylum-level classification for each MAG as predicted by GTDB-tk. Above this, the ‘References’ coloured ring indicates the phylum-level classification of the bacterial reference sequences. The outer ring bar plot shows the number of PHA depolymerases predicted in each MAG. The type(s) of predicted PHA depolymerase(s) (mcl, sclT1, or sclT2) found in that MAG are indicated with coloured squares on the innermost rings.
Reference ePHAmcl depolymerases originated from the Bdellovibrionota, Gammaproteobacteria and Actinobacteriota. The study identified PHAmcl depolymerase-encoding MAGs from the Proteobacteria, Firmicutes and Myxococcota, with the highest overall frequency occurring in the Firmicutes (Figure 4).
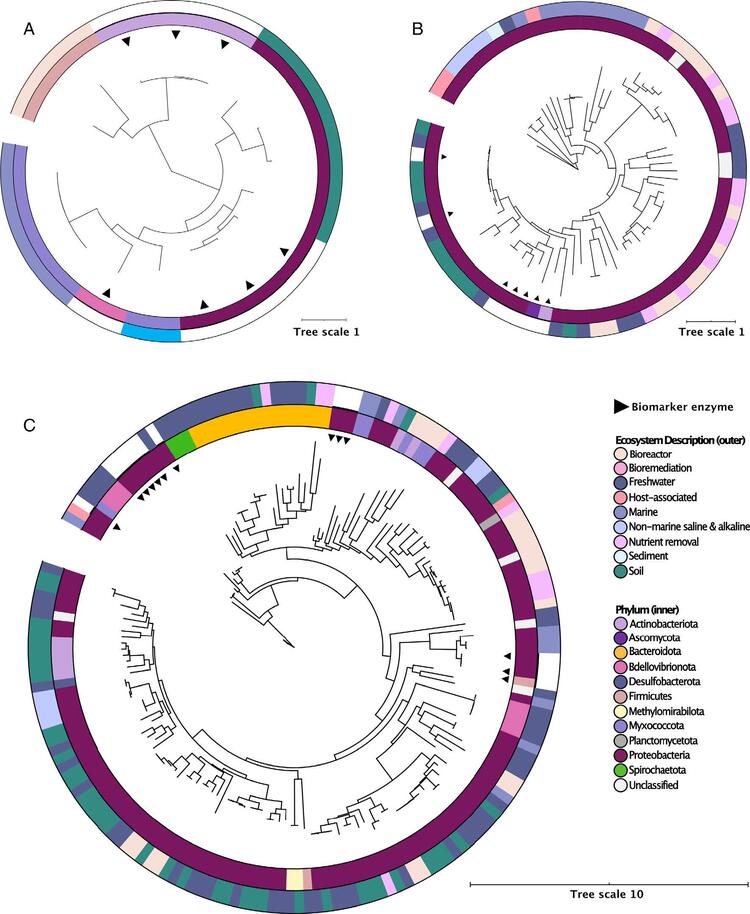
Figure 4: Environmental origin and phylogenetic relationships of the PHA depolymerases predicted from all MAGs. Tree scale bar indicates number of substitutions per site. Outer rings are coloured by environmental origin of predicted PHA depolymerases. Inner rings are coloured by phylum of the MAG encoding the predicted PHA depolymerase. ML trees showing (A) 10 (eight unique) PHAmcl depolymerases (B) 156 (133 unique) PHAscl T1 depolymerases and (C) 65 (56 unique) PHAscl T2 depolymerases
Conclusions
Biodegradable plastics such as PHA offer a promising solution to addressing the significant environmental effects caused by conventional plastics. As PHA degradation is largely reliant on the abundance and diversity of microorganisms encoding PHA depolymerases, it is important to understand their distribution and diversity.
This study presents a carefully curated set of predicted PHA depolymerases to conservatively catalogue the existing environmental sequence diversity and the associated taxonomic affiliations of this enzyme family. It significantly expands the inventory of PHA depolymerases available to target for future biochemical characterization and affiliates many of these enzymes with novel taxonomic lineages for this function. It also provides an overview of the occurrence and frequency of PHA depolymerases detected in the environment. Future research should employ a combination of metagenomic predictions, burial degradation trials with functional profiling and biochemical validation of predicted enzymes (particularly for enzymes distantly related to biomarker enzymes) in targeted environments to confirm predictions developed here.
Viljakainen, V.R. and Hug, L.A. (2021). The phylogenetic and global distribution of bacterial polyhydroxyalkanoate bioplastic-degrading genes. Environmental Microbiology. https://doi.org/10.1111/1462-2920.15409
Image by Hamsterfreund from Pixabay
For more information about WaterResearch, contact Julie Grant.






