It is estimated that anaerobic oxidation of methane (AOM) lowers net global emissions of methane by 10 to 60%, significantly mitigating the potential impact of this potent greenhouse gas on the global climate. Although certain microorganisms are known to carry out AOM alone, such as Candidatus Methylomirabilis oxyfera, AOM can also result from syntrophic (i.e. “eating together”) microbial interactions. Specifically, anaerobic methanotrophic archaea (ANME) perform AOM in association with sulfate-reducing bacteria (e.g. Desulfosarcina and Desulfococcus) or nitrite/nitrate-reducing bacteria (e.g. Candidatus Methylomirabilis oxyfera of the NC10 division and Candidatus Methanoperedens nitroreducens of the ANME-2d) Thus sulfate, nitrite, or nitrate can serve as electron acceptors for AOM.
The physiological mechanisms underpinning AOM are not fully understood, partly due to the lack of pure cultures. ANME-enriched cultures have certain characteristics in common with methanogens, such as the lipid structures and the presence of methyl-coenzyme M reductase. Available evidence indicates that reverse methanogenesis is one of the main pathways of AOM by ANME. In addition to ANME, known methanogens are also able to catalyze AOM, including pure cultures of Methanobacterium ruminantium, Methanobacterium strain M.o.H., Methanosarcina barkeri, and Methanospirillum hungatii.
Considering the abundance of iron- and manganese-oxide solids in methane-rich subsurface environments, metal-associated AOM may occur naturally in a manner similar to sulfate- and nitrite-dependent AOM, and the energetics appear to be more favourable. Coupling the reduction of metal oxides with AOM implies that AOM is associated with extracellular electron transfer (EET), which is necessary for reducing solid electron acceptors. Recent publications have suggested that EET is potentially involved in AOM between ANME and sulfate-reducing bacteria, where EET is unnecessary since sulfate is a soluble terminal electron acceptor. Although EET is indispensable for using solid forms as a terminal electron acceptor, previous studies have not focused on EET coupled to AOM using metal solids or electrodes as the terminal electron sink. In this study, we provide the direct experimental evidence that AOM can be coupled to EET by using electrodes as the terminal electron sink in a gas-tight microbial electrochemical cell (MxC).
Methodology
Dual-chamber MxCs were fabricated with Plexiglas (Figures 1 and 2). The working volumes of the anode and the cathode chambers were 280 mL and 122 mL, respectively. Carbon fibers were used as the anode, combined with a stainless steel current collector. Stainless steel mesh was used as the cathode. The two chambers were separated by an anion exchange membrane. Return activated sludge from the Waterloo Wastewater Treatment Plant was inoculated into the MxC anode chamber. The anode potential was fixed at -0.4 V versus a reference electrode. With cascade operation of the MxC with acetate medium and methane/acetate medium in batch mode, we operated the MxC solely with methane medium for over 300 days.
In order to characterize the reactor microbial communities, we collected biofilm samples at the end of the experiments. The top part of the combination valve from a Tedlar gas sampling bag was used as the gas sampling port. Genomic DNA was extracted, sequenced, and reads were then uploaded to the MG-RAST server for annotation. Archaeal and bacterial taxa were assigned using best-hit classification and functions of AOM were annotated by hierarchical classification based on the KEGG Orthology database. The AOM-related nucleotide sequences were selected and downloaded from the hierarchical classification results. Protein sequences for construction of AOM databases were extracted from Genbank. The BLASTx analysis was performed with standalone BLAST software with the selected nucleotide sequences and the constructed databases of AOM.
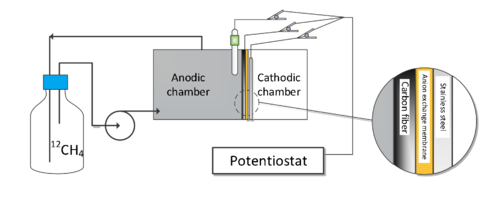
Figure 1. Schematic of a gas-recirculation loop system for the microbial electrochemical cell operated inside an anaerobic chamber (MxCAC).

Figure 2. Photo of the MxCAC equipped with a loop system inside an anaerobic chamber.
Outcomes
The results demonstrate that AOM was coupled with EET to the anode, generating electric current. The net electric current from AOM ranged from 6.6 to 13.6 mA/m2 (average, 11.0 ± 1.3 mA/m2) in the MxC when non-Faradaic current is taken into account for the abiotic electrochemical cell.
The majority of archaeal small subunit rRNA gene sequences corresponded to Methanobacterium (67%). In comparison, Geobacter (38%) dominated the bacterial small subunit rRNA gene sequences. From functional annotation, most of the genes essential for methanogenesis also have been identified in AOM were detected and represented 24% of the reads annotated to methane metabolism. According to taxonomic identification, the AOM-pathway genes belonged to the family Methanobacteriaceae, including Methanobacterium and Methanothermobacter, but none of these genes were associated with the Geobacter genus. Considering the results of small subunit rRNA gene data, most AOM genes were affiliated with Methanobacterium-related archaea. Cytochrome c and type IV pili are associated with EET and were found in the metagenome at 0.96% and 0.16% of total functional abundance, respectively. Geobacter-related bacteria accounted for 89% of c-type cytochrome annotations and 94% of type IV pili annotations. In contrast, Methanobacterium spp. did not contribute any of these genes to the metagenome.
Microscopic analysis demonstrated that members of the Methanobacteriaceae family and Geobacter genus co-existed on the surface of the anode’s carbon fibers, supporting the hypothesized syntrophy between Methanobacterium spp. and Geobacter spp. for enabling the EET-coupled AOM reaction using the anode as a terminal electron acceptor, not direct electron transfer by Methanobacterium spp. alone (Figure 3). Figure 3a) shows the 3D structure of the colonies formed by Methanobacteriaceae family (green) and Geobacter genus (red) on the surfaces of a single carbon fiber. Yellow indicates the overlap of green and red colors. Figure 3b) shows biofilm structure in 2D. Image 3a) is obtained from a confocal microscope, and image 3b) is from an epifluorescence microscope.
We did not find multi-heme cytochromes in the Methanobacterium genus. Instead, the metagenome in the study was affiliated with formate dehydrogenase and hydrogenase genes, and over 50% of these genes were annotated as being associated with Geobacter-related bacteria. The reads of formate dehydrogenase and hydrogenase genes imply that Geobacter spp. might oxidize formate or hydrogen generated from AOM catalyzed by Methanobacterium (i.e. reverse methanogenesis). This suggests that the consortium of Methanobacterium and Geobacter might employ intermediate-dependent interspecies electron transfer.

Figure 3. Fluorescence in situ hybridization images of intact biofilms on carbon fibers from the MxCAC anode.
Conclusions
Our study documents EET-coupled AOM in a biofilm anode, probably via intermediate-dependent inter-species electron transfer between Methanobacterium and Geobacter. This newly discovered syntrophic interaction implies that certain methanogens can work synergistically with bacteria for respiring electrons to metal-oxide solids. This syntrophy is unexpected because methanogens are viewed conventionally as competitors with bacteria that reduce metal oxides, competing for available electron donors. Although unexpected, this new syntrophy could have substantial implications for lowering estimates of global methane flux, because methanogens and metal-oxide solids are widely distributed in sedimentary and terrestrial environments.
Gao, Y., J. Lee, J.D. Neufeld, J. Park, B. E. Rittmann, and H. Lee (2017). Anaerobic oxidation of methane coupled with extracellular electron transfer to electrodes. Scientific Reports 7, 5099.
Contact: Hyung-Sool Lee, Civil and Environmental Engineering, Jangho Lee, Department of Civil and Environmental Engineering; Josh Neufeld, Department of Biology
For more information about the Water Institute, contact Amy Geddes.







