Background
Relations and functions in living systems are complex and entwining due to intertwining unknown factors of subjects and environment. Currently, protein binding predictions are based on residue-residue close contact (R2R-C) data procured from existing 3-dimensional protein-protein interaction complexes (Fig.1). However, numerous physio-chemical/stereo factors in the complex interacting environment might affect R2R-C (Fig.2). R2R-C association patterns could be masked at the data level and misleading, yielding unreliable Residue to Residue Interaction (R2R-I) predictions. Therefore, new methods capable of discovering and disentangling R2R-I patterns are needed to enhance the understanding and prediction of protein-to-protein interactions with reasons rather than just giving a black-box decision.
Description of the invention
University of Waterloo (UW) researchers have developed a novel software P2K (Pattern to Knowledge) to discover deep knowledge inherent in bio-sequences such as DNA/RNA/Protein/Antibody. P2K uses novel autonomous and scalable algorithms to discover deep knowledge to reveal subtle functionality and relations and uses the knowledge discovered to enhance machine learning, achieving much better prediction results with explanation. The developed software receives two bio-sequences as the input and outputs the anticipated binding sites between the two.
Advantages
- Time/Cost reduction - uses sequence data only with no experiment setup or reliance on explicit prior knowledge.
- Higher accuracy – enhanced ML model by extracting predictive features from Protein to Protein-Interaction sequence data from the cloud.
- Robust to data noises - removes data noise and minimizes misinformation from entangled factors/sources.
- Flexible - applicable to other scenarios such as protein to antibody and protein to aptamer.
- Experimental comparison: a) 28% better prediction than existing solutions b) capable of revealing and confirming binding sites of interacting proteins not achievable by other methods.
Potential applications
- Assist in drug development
- Genomic, transcriptomic and proteomic data analysis
- Gene therapy – precision medicine
- Environmental detection (e.g., Aptamers, DNA barcodes)
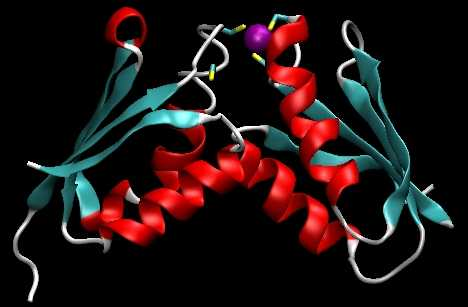
Fig. 1: Residue interaction between proteins
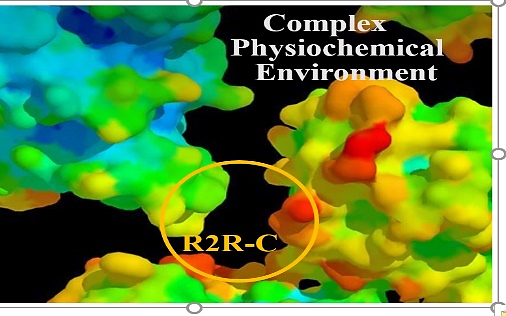
Fig. 2: Residue contact environment
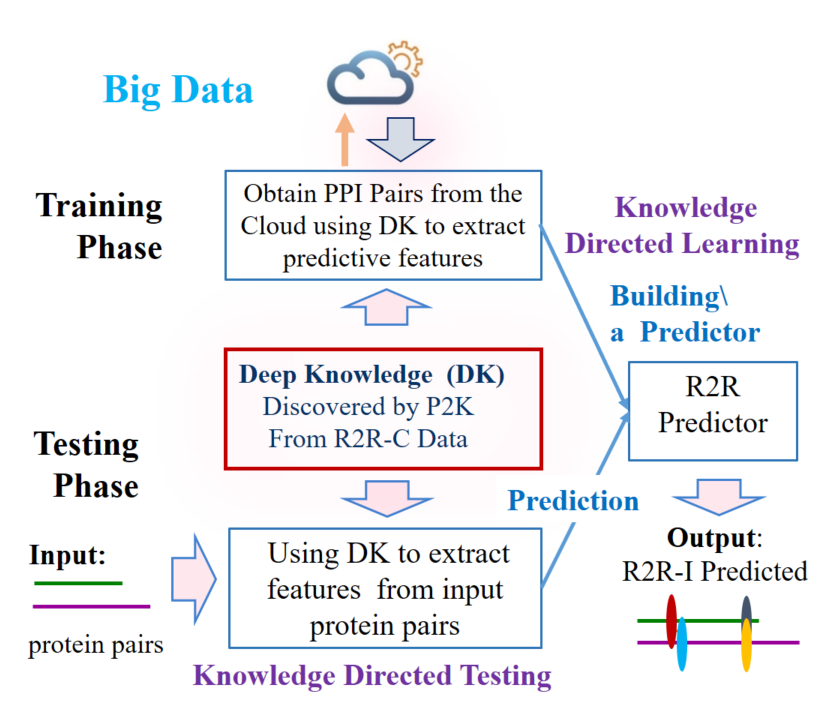
Fig. 3: Deep knowledge directed ML for R2R-I predictions
References
7423
Patent Status
US 11,074,992
Stage of development
Working prototype and validating application data
Contact
Scott Inwood
Director of Commercialization
Waterloo Commercialization Office
519-888-4567, ext. 43728
sinwood@uwaterloo.ca
uwaterloo.ca/research