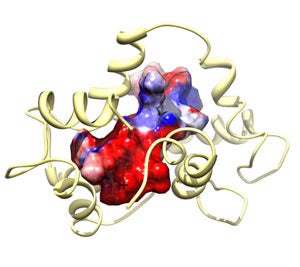
Structure of Calmodulin (CaM) Bound to Endothelial Nitric Oxide Synthase Peptide phosphorylated at Thr 495 shows the spacefilling model of the target peptide with the surface mapped by electrosdtatic charge (red = negative and blue = positive).
- structure: Piazza, M. et al Biochemistry 53(2012): 1241-1249.
- coordinates: PDB/RCSB entry 2MG5
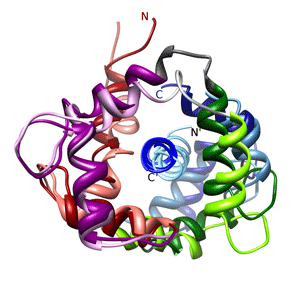
Structure of Calmodulin (CaM) Bound to Nitric Oxide Synthase Peptides shows the iNOS and eNOS target peptides in complex with calmodulin superimposed on the N-lobe. The iNOS complex is shown in the more intense colors whereas the eNOS complex is shown in pale colors.
- structure: Piazza, M. et al Biochemistry 51(2012): 3651-3661.
- coordinates: PDB/RCSB entry 2LL6
- and PDB/RCSB entry 2LL7
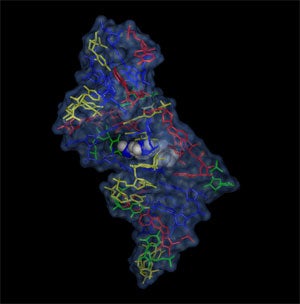
The malachite green - RNA aptamer complex shows the ligand deeply embedded in the core of the RNA. The complex is stabilized by electrostatic forces and stacking interactions. Adenines are shown in red, guanines in blue, cytosin in yellow and uridines in green.
- structure: Flinders, J. et al ChemBioChem 5(2004): 62-72.
- coordinates: PDB/RCSB entry 1Q8N
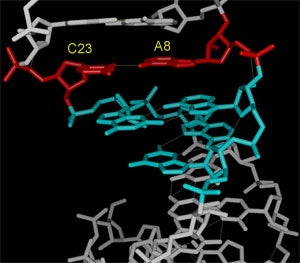
Substrate stem-loop of the VS ribozyme. The structure illustrates how the internal loop around the cleavage site forms a completely base paired bulge that contains tandem G:A base pairs and an A+:C base pair (the A is protonated). Further studies of the structure revealed a pH dependent conformational switch that causes the A in the A+:C pair to loop out of the helix at high pH.
- structure: Flinders, J. et al J. Mol.Biol. 308(2001): 665-679.
- coordinates: PDB/RCSB entry 1HWQ
The active site loop (Loop VI) of the VS ribozyme contains the nucleotides that are most likely involved in the chemical catalysis step. The structure of the loop shows a high degree of dynamics within the RNA backbone. We assume that this is related to the transition between active and inactive conformations of the ribozyme.
- structure: Flinders, J. et al J. Mol. Biol. 341(2004): 935-949.
- coordinates: PDB/RCSB entry 1TJZ

Molecular graphics on this page were generated using the programs MOLMOL and UCSF Chimera in conjunction with the Persistence of Vision (POV-Ray3) ray tracing package.