
BIOGRAPHY
My name is Brenda and I have been in Waterloo my entire life. I was born and raised here and have attended UofWaterloo since 2009. I completed my BSc in Biomedical Science with a Physics Minor in 2012 and finished my PhD in Biology (Nanotechnology) in 2017 under the supervision of Professor Zoya Leonenko. During this time, I was awarded a Waterloo Institute of Nanotechnology Nanofellowship. Since the completion of my doctoral studies, I have continued to work as a Research Associate in the Nanoscale Biophysics Group. I am currently a full-time Lecturer and Life Physics Academic Advisor in the Department of Physics & Astronomy, while still volunteering as a Research Associate to continue my research interests.
I have a passion for teaching and received the 2014 Amit & Meena Chakma Award for Exceptional Teaching by a Student and 2019 WUSA Teaching Award. I also received the 2015 SWAAC Graduate Award for outstanding leadership as President of Tutoring Beyond Borders, a charity I started to provide free tutoring to underprivileged, local high school students.
EDUCATION
2009 - 2012: BSc Honours Biomedical Sciences, Physics Minor
2013 - 2017: PhD in Biology (Nanotechnology)
2017 - 2019: Research Associate
2019 - Current: Lecturer, Volunteer Research Associate
RESEARCH
The world we live in is constantly adapting to changes that arise from scientific breakthroughs and technological innovations. What’s even more fascinating is that the same phenomena occur on the nanoscale within the microbial world. As humans discover and use new drugs and medications, the bacteria that these antibiotics aim to destroy can adapt, mutate their genes, and evolve to thrive in these new conditions. Unfortunately, that means that these bacteria are now antibiotic-resistant and it’s back to the drawing board for humans to try and come up with new antibiotics to target the same microbe.
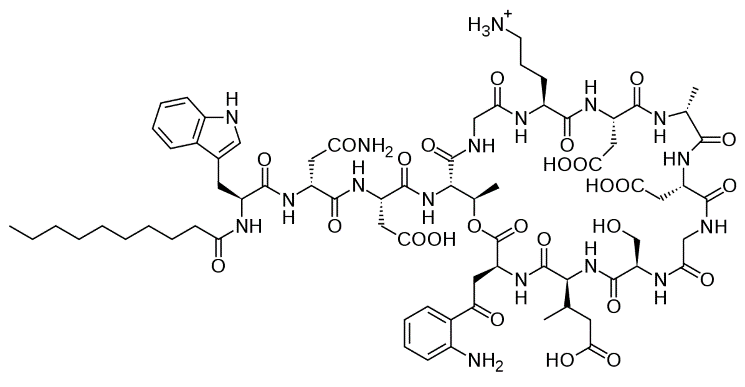
Figure 1. Chemical structure of daptomycin
My biophysics research focuses on an antibiotic called daptomycin. But what’s so special about this drug? First of all, it’s an antimicrobial peptide, an antibiotic that is naturally-derived from a living organism. In this case, daptomycin is derived from the actinobacteria Streptomyces roseosporus. Second, daptomycin is the only member of the cyclic lipopeptide class of antibiotics. You may picture the structure of daptomycin as a circular head with a long tail (Figure 1). Third, daptomycin has a distinct mechanism of action due to its unique structure: it has been proposed that its tail inserts into the bacterial membrane and when a group of daptomycin molecules come together, a pore is formed that leaks ions and causes a destructive loss of membrane potential that kills the bacterial cell (Figure 2). This unique mechanism of action has made daptomycin an extremely useful asset in treating infections caused by drug-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE). In fact, daptomycin has been proven to be effective against all clinically-relevant Gram-positive bacteria and its bacterial resistance is very low.
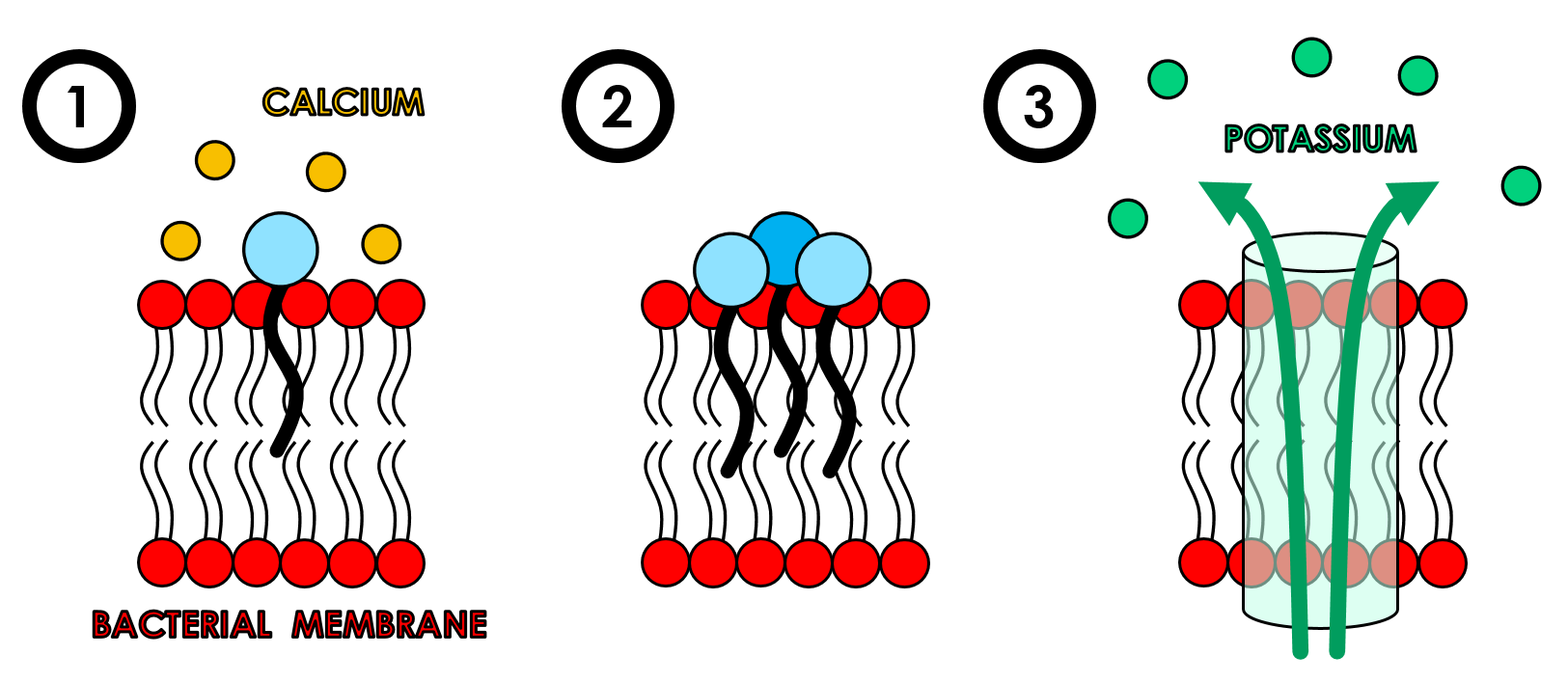
Figure 2. Proposed mechanism of action for daptomycin. 1) Daptomycin’s tail binds to and inserts into the bacterial membrane in the presence of calcium ions. 2) Daptomycin groups together to form aggregated structures. 3) These aggregates form ion channels, or pores, at which point intracellular potassium is released, membrane potential is disrupted and therefore causes cell death.
Despite daptomycin’s unique traits, there is one major problem. Theoretically, daptomycin should be able to exhibit bactericidal activity against Gram-positive Streptococcus pneumoniae in cases of community-acquired pneumonia. However, the pulmonary surfactant that lines the lung’s alveoli somehow inhibits daptomycin, making it the first case of organ-specific antimicrobial peptide inhibition. Considering S. pneumoniae has grown resistant to treatments by penicillin and amoxicillin, it is therefore imperative that we try and elucidate daptomycin’s mechanism of action and its hindrance by pulmonary surfactant. As a result, my research goals are to shed more light into how daptomycin acts on bacterial cells and why daptomycin is ineffective against S. pneumoniae cells inside the lungs.
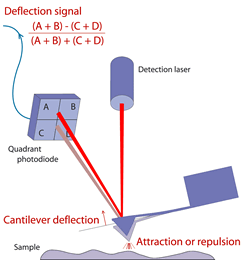
Figure 3. Schematic of atomic force microscopy. The quadrant photodiode detects deflections from the cantilever as it scans across the sample surface, interacting through attractive or repulsive forces. Image courtesy of JPK Instruments AG.
Having a background in both Biology and Physics, working in Dr. Zoya Leonenko’s Nanoscale Biophysics Group is the perfect marriage for both my passions. I get to use methods and theories from physics to study a biological issue on the nanoscale. To study the effect of daptomycin on bacterial membrane and lung surfactant lipid models, I use atomic force microscopy (Figure 3) or AFM to observe any structural changes that take place in the presence of daptomycin by taking images of the sample’s surface. To understand how AFM works, imagine that the AFM has a tip that is comparable to the needle of a record player and that the sample surface is like the vinyl record. This tip runs back and forth along the sample surface, experiencing attractive and repulsive forces across each area it touches while sending feedback signals to the detector so that a topographical image is created on the computer’s software. I also use Kelvin probe force microscopy, which is similar to AFM, but it measures local charging in different areas of a conductive sample. Apart from imaging methods, fluorescence spectroscopy and the Langmuir-Blodgett trough technique are used to study the insertion of daptomycin between the different lipid models. This research will help us further our understanding of daptomycin’s mechanism of action and steer future development and optimization of daptomycin derivatives to combat S. pneumoniae inside the lungs.
RESEARCH INTERESTS
- Study of antimicrobial peptide daptomycin and its interaction with pulmonary surfactant and model bacterial membranes
- Use of atomic force microscopy, Kelvin probe force microscopy, Langmuir-Blodgett trough, fluorescence spectroscopy and black lipid membrane techniques
LEONENKO LAB HISTORY
- May 2011 - Aug 2011: NSERC USRA Summer Student (Physics)
* Working with atomic force microscopy imaging of amyloid-beta interactions with SG1 inhibitors - Sep 2011 - Dec 2012: Undergraduate Research Volunteer (Biology)
* Atomic force microscopy imaging of DHA (omega-3) and its effect on amyloid-beta fibril formation - Jan 2013 - Apr 2017: PhD Candidate in Biology (Nanotechnology)
- May 2017 - Current: Research Associate