ESRP1 Role in GI Development
Knockout of ESRP1 is perinatal lethal in mice, and simultaneous ablation of ESRP1 and paralogue ESRP2 is perinatal lethal in zebrafish. ESRP1 knockdown and knockout results in widespread transcriptomic changes, most well studied is the alternative splicing changes. Knockout of ESRP1 alone results in several dysmorphic phenotypes including Cleft Lip/Cleft Palate, inner ear defects and kidney branching defects. ESRP1 loss also contributes to several to the failure to develop several many organs in the mice, including salivary glands, lungs, and colon. Reduction in wildtype ESRP1 is sufficient to compromise colonic barrier integrity and contribute to colon shortening. Our lab seeks to understand how ESRP1 regulates GI function using 3D models of colon development.
Investigating ESRP1
Binding and Determining How ESRP1 Targets are Identified and Defining the Roles of ESRP1 Binding in the UTR of Genes
Transcriptome-wide mapping of ESRP1 binding shows enriched binding in the 3’UTR of protein coding genes. ESRP1 has well established roles in regulating alternative splicing, however, the function of ESRP1 in the UTR is unknown. While several studies have alluded to roles of ESRP1 in regulating mRNA translation the mechanism is undefined. Furthermore, while we have identified the binding motif of ESRP1 in vivo, it remains unclear how specific ESRP1 sites are chosen. It remains to be elucidated what are the trans-acting and cis-regulatory elements that determine the context of ESRP1 of ESRP1 binding in vivo. Our lab seeks to understand ESRP1 interaction with its targets using in vitro and cell-based models.
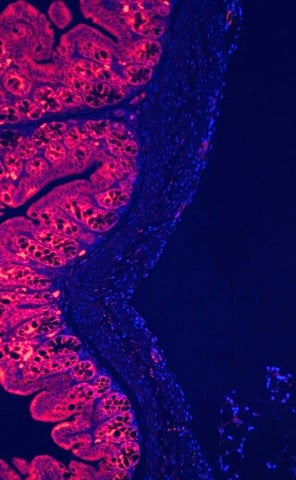
Immunofluorescent image of the mouse intestines showing expression of ESRP1 protein in the intestinal epithelial cells. Stained and imaged by SK Lee.

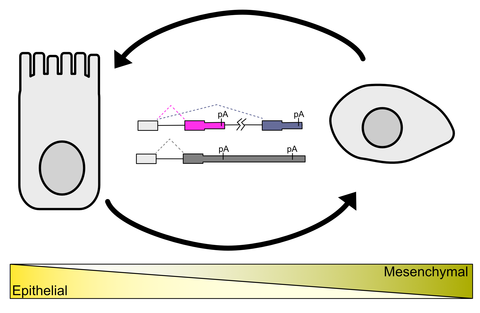
Alternative Polyadenylation
And its Impact on Epithelial Integrity and Mosaicism in Epithelial to Mesenchymal Transition
Cleavage and polyadenylation is a highly regulated constitutive process required for generation of mature mRNA. However, differential recruitment and association of the regulators of polyadenylation can lead to tissue specific and disease specific alternative polyadenylation (APA) which may alter the length of the 3’UTR of the mRNA or generate truncated mRNA transcripts. This results in the generation of different mRNA isoforms that may have differences in their biological functions. Dysregulation of APA is a feature of many cancers, and several APA regulatory proteins have been implicated with cancer progression. We aim to investigate the impact on APA on epithelial cell integrity and to investigate the mechanisms by which APA contributes to epithelial to mesenchymal transition.