The main protease (M pro ) in severe acute respiratory syndrome coronavirus 2 (SARS- CoV-2), the virus responsible for the coronavirus disease (COVID-19), has emerged as a promising drug target. The scientific community has produced a large number of crystallographic structures of the protease, which mediates viral replication and transcription. These structures report several fragments with varied chemotypes binding to different sites in M pro . The main challenge at this stage is to effectively corroborate these valuable structural insights and expedite the search for any known drugs or natural products with properties similar to those fragments such that they can be rapidly translated for clinical testing against SARS-CoV-2. In this project, we build an artificial intelligence-based model using the available structural data of fragment-bound SARS-CoV-2 M pro complexes.Leveraging known drug-target interactions, our goal is to produce a machine learning algorithm capable of predicting potential drugs that can be repurposed for the treatment of COVID-19. We explore the potentials of small molecules, including drugs, natural products, and quantum dots to identify promising structures for inhibiting and/or detecting the SARS-CoV2 virus. Our results are expected to provide useful insights into the readily available therapeutic resources and help in the fight against the COVID-19 pandemic.
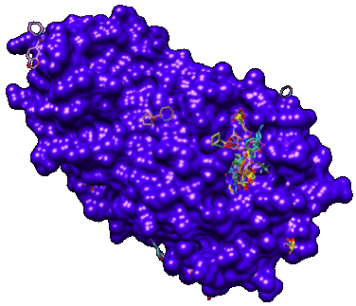
Figure 1. The X-ray crystal structures of the recently reported fragment-bound SARS-CoV-2 Mpro complexes from Protein Data Bank. The structure of Mpro is shown as a blue surface, and all the small fragments are represented as stick representation.