Background
Identifying functional segments or regions from bio-sequences is a major challenge in bioinformatics. Functional segments of a bio-sequence could reveal folded structure, physio-chemical functionality and mutation hotspots for better understanding of biological mechanisms; directing the design of new drugs and discovering new knowledge about the cure of genetic diseases. With explosive data streaming in, effective, accurate and scalable methods are still lacking. Existing methods such as: MEME, GLAM2 are incompetent to capture frameshift and rare mutations.
Description of the invention
University of Waterloo (UW) researchers have developed a novel software that uses a systematic process to align pattern clusters of bio-sequence families and thereby, to identify functional regions. The software also adaptively determines the width and mutation spots without relying on exhaustive search and without relying on explicit prior knowledge or clue. While the software discovers new patterns with strong statistical support, it also spots mutational rare patterns with minor substitution and frameshift (insertion and deletion). This is of ample importance for personalized medicine, gene therapy/marker and drug research.
Advantages
- Allows variable pattern length
- Capable of identifying mutations and rare mutations (Fig. 2)
- Fast (400X compared to MEME method), accurate, and precise (location-wise)
- Does not need parameter pruning (compared to MEME, GLAM2 method)
- No explicit prior knowledge needed
- Compatible with hardware acceleration/multitasking
From the APCs discovered, the software can disentangle patterns within APCs to further reveal deeper knowledge on subgroup characteristics in different specific statistical/functional spaces with/without class labels given.
Potential applications
PD-APCn software can be used in drug discovery, personalized medicine and gene therapy/marker for:
- Bio-sequence functional region identification
- Residue-Residue and Protein-Protein interaction prediction
- Protein-DNA binding cores discovery
- Drug-able site discovery
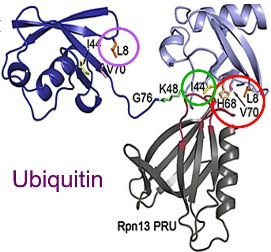
Fig. 1 APCs discovered (colored circles) contain key binding elements
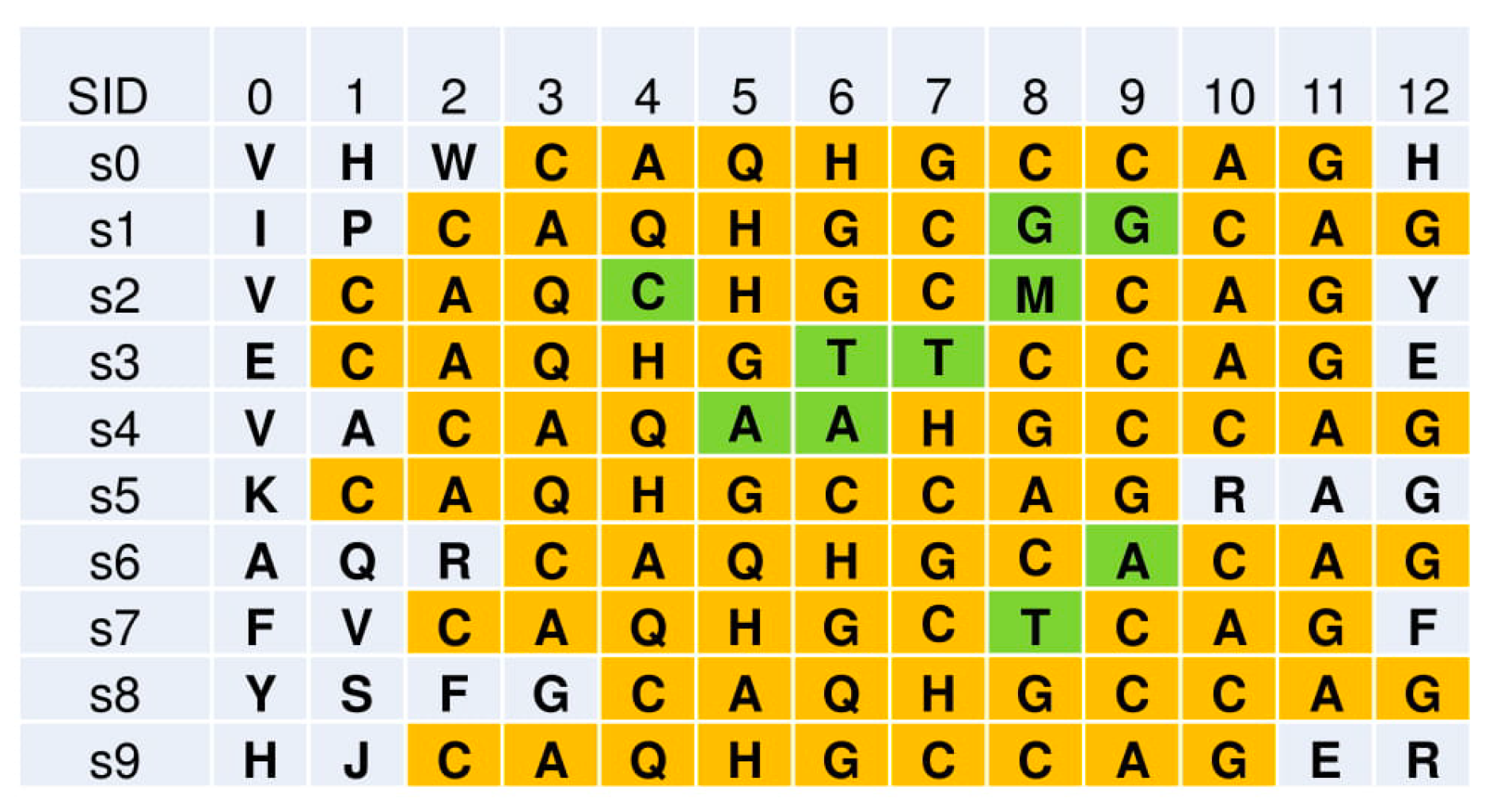
a. Input: Synthetic sequence data with implanted mutations
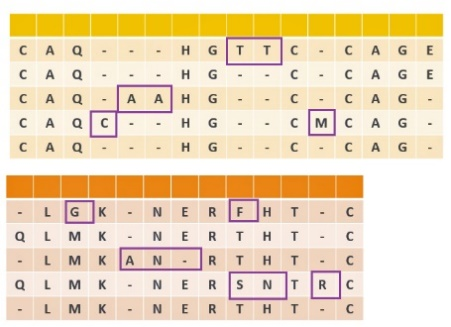
b. Output: all mutated patterns discovered in APCs
Fig. 2 Pattern-Directed Aligned Pattern Clustering
Reference
10145
Patent status
Patent Pending
Stage of Development
Working server prototype
Contact
Scott Inwood
Director of Commercialization
Waterloo Commercialization Office
519-888-4567, ext. 43728
sinwood@uwaterloo.ca
uwaterloo.ca/research