Extremophiles are species that are adapted to live at the edges of biological tolerance, in a range of environments that seem inhospitable to life by human standards. These extremely hardy organisms are found in all three domains and all six kingdoms of life, the highest and second highest levels of classification biologists use to categorize living things based on common ancestry.
For example, tardigrades, tiny eight-legged invertebrates in the animal kingdom, are found in harsh environments as diverse as deserts, mountain tops, glaciers and hot springs. Similarly, among organisms in the plant kingdom, a saltwater cress species known taxonomically as Thellungiella salsuginea thrives in cold, alkaline flats at the edges of saline pools in the Yukon.
But the extremophiles perhaps most deserving of the name are certain species of highly specialized bacteria and their distant relatives, the archaea — microbes so evolutionary different from each other that they are in different domains of life.
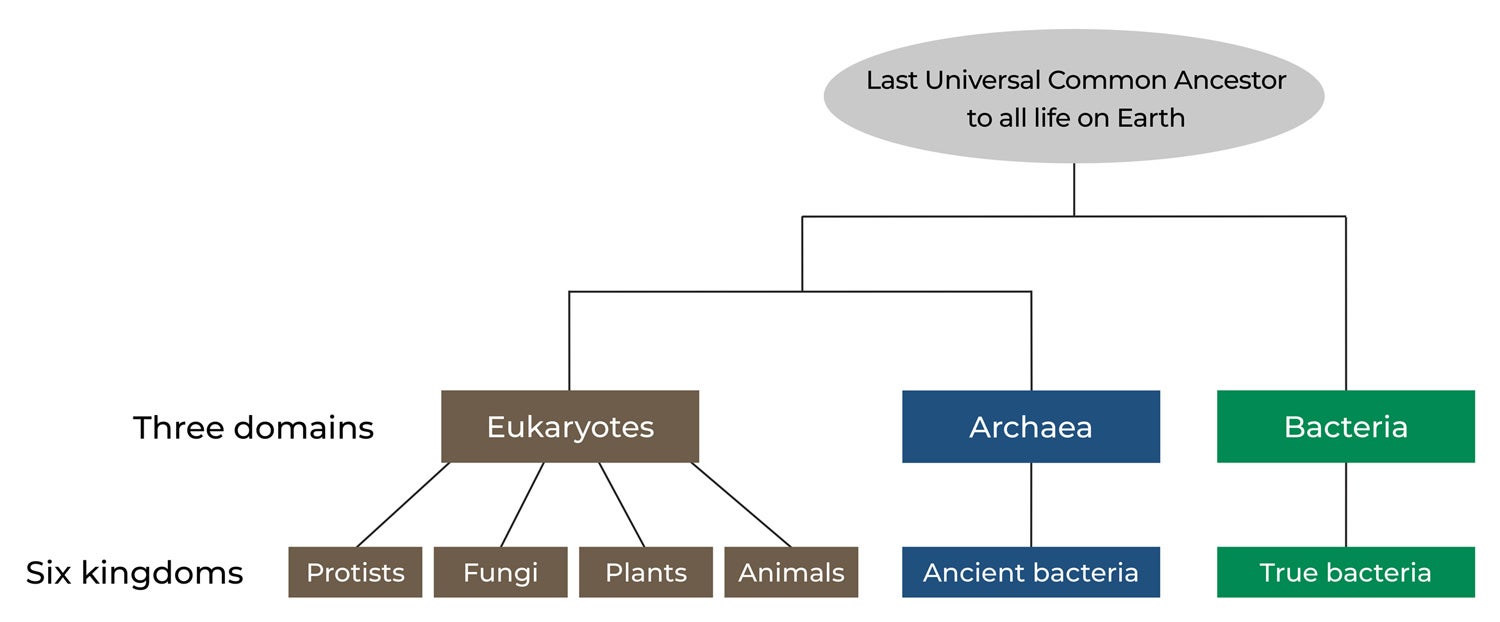
The Tree of Life has three domains — bacteria, archaea and eukaryotes — and six kingdoms. All life on Earth belongs to a given domain and a kingdom within it. The Last Universal Common Ancestor is a hypothetical common ancestral cell that lived some four billion years ago from which the three domains of life are thought to have originated.
For that reason, you would not expect bacterial and archaean extremophiles adapted to similar extreme environments to have similar genomic signatures. But according to a new study in Nature Scientific Reports by Cheriton School of Computer Science researchers and their colleagues at Western University and the University of PEI, adaptations to extreme temperatures and to extreme pH imprint a discernible environmental component into the genomic signature of microbial extremophiles.
The team evaluated a dataset of 700 microbial extremophile genomes using machine learning algorithms that looked for genomic similarities across them.
“Our study detected a strong environmental signal in the genomes of unrelated extremophiles that live in similar extreme environments,” said Cheriton School of Computer Science Professor Lila Kari. “At first, we could not believe our eyes, as it was so unexpected. This is in a way akin to finding out that your human DNA is more similar to the plant DNA of flowers in your garden, than to the human DNA of your cousin who lives on another continent.”
L
to
R:
Professor
Lila
Kari
and
PhD
student
Pablo
Millán
Arias,
the
authors
of
the
study
from
the
University
of
Waterloo.
(Study
collaborators
Joseph
Butler,
Maximillian
Soltysiak,
and
Professor
Kathleen
Hill
from
the
University
of
Western
Ontario,
and
Gurjit
Randhawa
from
the
University
of
Prince
Edward
Island,
were
unavailable
for
the
photo.)
Pablo
Millán
Arias’s
research
interests
span
from
deep
learning
for
DNA
classification
and
clustering
to
information
theory
and
information
geometry.
Lila
Kari
is
a
Professor
in
the
Cheriton
School
of
Computer
Science.
Author
of
more
than
200
peer-reviewed
articles,
she
is
regarded
as
one
of
the
world’s
experts
in
biomolecular
computation.
To understand how the research team detected an environmental signal in the genome of bacterial and archaeal extremophiles requires a digression into the structure of DNA, mathematical ways to represent DNA sequences, and different kinds of machine learning techniques used for taxonomic classification and identification.
In the same way that we use letters of the alphabet to write words and sentences, and bits 0 and 1 to write computer machine code, the four basic DNA units — the nucleotides adenine (A), cytosine (C), guanine (G), thymine (T) — are used by nature to write genetic information as DNA strands.
This sequence of nucleotides in a fragment of DNA extracted from the genome of an organism can be analyzed mathematically in a number of ways. The researchers first generated genomic signatures by calculating, for a given number k, the number of times each string of length k (called k-mer) occurs in the DNA fragment. For example, if the nucleotide sequence A-G-T-C-G occurred 20 times in a DNA fragment, this particular 5-mer would have a count of 20. From this k-mer data, a numerical vector of k-mer counts can be constructed for each DNA fragment, and it becomes the genomic signature unique to that genome and organism.
These genomic signatures are then used to train a supervised machine learning model, Professor Kari explains. “We give a supervised machine learning algorithm a genomic signature and its taxonomic label — in other words, we tell the algorithm that this particular genomic signature is from a bacteria species, and this genomic signature is from an archaea species. If you train the algorithm with many genomic signatures that are taxonomically labelled, it will learn from these examples and become able to classify new genomic sequences as bacteria or archaea with very high accuracy, as high as 95 percent.”
The twist is that the research team then did the same thing, except instead of giving the supervised model the genomic signatures with their corresponding taxonomic labels, they gave the algorithm genomic signatures with their environmental information labels.
“We told the model that this microbe is a hyperthermophile, this microbe is a thermophile, this microbe is a psychrophile, a kind of extremophile that thrives at low temperatures,” Professor Kari said.
“It’s the exact same genomic data set as before, except we told it nothing about the organism’s taxonomy, only what kind of extreme environment it lives in,” she continued. “When we trained the model with environmental labels and asked it to predict the environment corresponding to the DNA fragment from a new organism, we still got accurate classification. That’s the truly surprising result. If all of the information contained in the genomic signature were strictly taxonomic, as previously believed, we should not be able to predict the environmental conditions that extremophiles live in based on their genomic DNA fragments. Granted, the environmental signal was somewhat weaker than the taxonomic signal, but the classification accuracy by environment type was still high, over 80 percent.”
A problem in using supervised machine learning to determine taxonomy, however, is that an estimated 95% of forms of life on Earth have not yet been catalogued and classified.
“We know so little about lifeforms on Earth that the chances are that if you present an unknown species to a supervised machine learning algorithm that has been trained with known species, it’s almost certainly guaranteed to get the wrong answer,” Professor Kari said. “To circumvent this limitation of supervised machine learning — that it can only recognize something that it has seen during training — and to check that the environmental signal we found was not a fluke, we then used an unsupervised machine learning approach.”
The same dataset of genomic signatures that was used earlier was given to the unsupervised machine learning algorithms, ones that did not know anything about either the organism’s taxonomy or its environment. Looking at the genomic signatures, the unsupervised machine learning model simply aimed to produce clusters of genomic signatures with similar genomic patterns. Surprisingly, some clusters that formed had both thermophilic bacteria and thermophilic archaea.
“We found some exemplars that always clustered together even though some were bacteria and others were archaea, microbes in different domains of life that are more vastly different evolutionarily from each other than a polar bear is from a mushroom.”
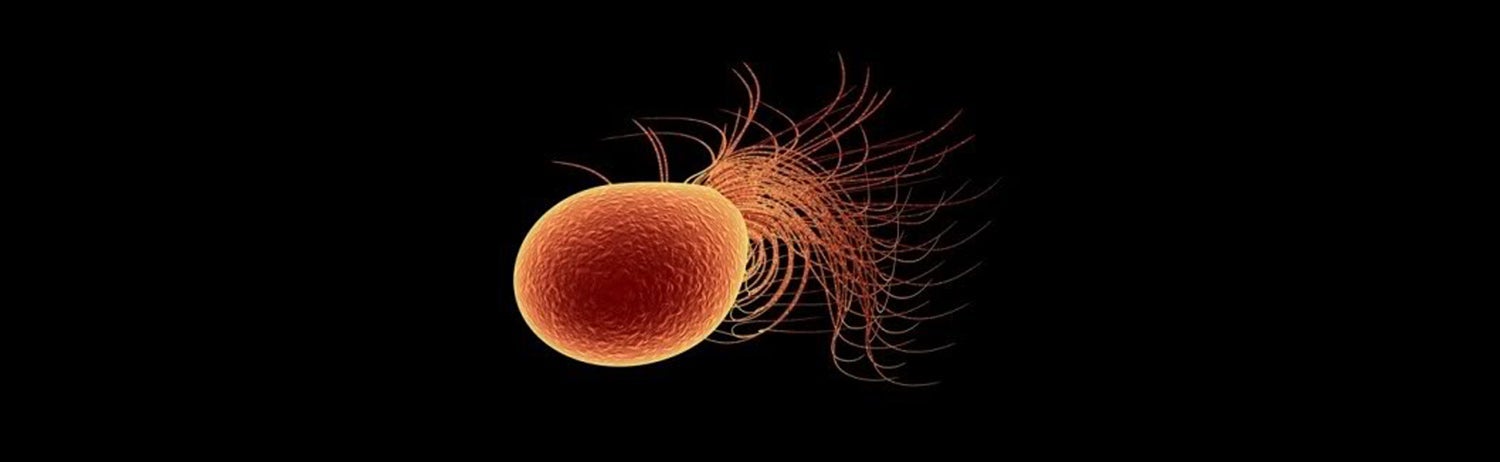
A
computer
rendering
of
the
archaeal
species
Pyrococcus
furiosus,
a
model
hyperthermophile
that
was
isolated
from
geothermally
heated
marine
sediments
at
the
beach
of
Porto
di
Levante,
Vulcano,
Italy.
Its
optimal
growth
is
at
100°C,
a
temperature
that
would
be
lethal
to
most
life.
Pyrococcus
furiosus
is
one
of
three
exemplars
of
hyperthermophilic
archaea in
the
study
whose
genomic
signature
was
grouped
together
as
similar
to
the
hyperthermophilic
bacterium,
Thermocrinis
ruber,
by
all
machine
learning
algorithms
used,
even
though
they
are
in
different
domains
of
life.
Art
made
by
Alfalo
provided
for
a
report
by
Michelle
Kropf.
Date:
2010.
Source:
https://en.wikipedia.org/wiki/Pyrococcus_furiosus#/media/File:P_furiosus.jpg.
CC
BY-SA
4.0
An obvious question is what could be the common factor that produced these clusters, as bacteria and archaea being clustered together cannot be explained by close ancestry. The only common trait found upon closer examination was that the clustered microbes were all thermophilic.
“This means that an environmental signature is present and indeed pervasive in the genome of certain extremophiles,” Professor Kari said. “That’s a fascinating insight, that an environmental signal exists, like a watermark, pervasive throughout the genome of some distantly related extremophiles that live in similar extreme environments, and that this environmental genomic signal can sometimes be stronger than even the genomic signal resulting from ancestry.”
Understanding what’s behind this environmental signal could uncover clues about the origin of life itself. Evolutionary studies of extremophiles strongly suggest that a hyperthermophile — an ancient microbe that lived about four billion years ago in hydrothermal vents on the ocean floor — was the universal ancestor of all life on Earth, the organism in the Tree of Life from which all current-day bacteria, archaea, and eukaryotes arose.
Studies of extremophiles have even farther-reaching implications, perhaps providing insight into the possibility of life being able to survive elsewhere. In a recent experiment, reported in the journal Frontiers in Microbiology, an extremophile bacteria species known as Deinococcus radiodurans, among the most radiation-tolerant organisms yet discovered, was transported from Earth to the International Space Station where it was exposed to the harsh conditions of space in an ultimate test of microbe survivability. Even after three years of exposure on a specially designed platform outside the International Space Station, Deinococcus radiodurans endured the vacuum, icy cold and intense radiation of space.
“Our study has revealed, in some sense, a new dimension of the genome: The DNA of extremophiles contains, in addition to ancestry information, information associated with the extreme environment where they live,” Professor Kari said. “This research could lay the groundwork for a ‘genomic observatory’ to explore and discover other such signals within the genome. The results could potentially lead to a deeper understanding of the history of life on Earth.”
To learn more about the research on which this feature article is based, please see Pablo Millán Arias, Joseph Butler, Gurjit S. Randhawa, Maximillian P. M. Soltysiak, Kathleen A. Hill, and Lila Kari. Environment and taxonomy shape the genomic signature of prokaryotic extremophiles. Sci Rep 13, 16105 (2023).